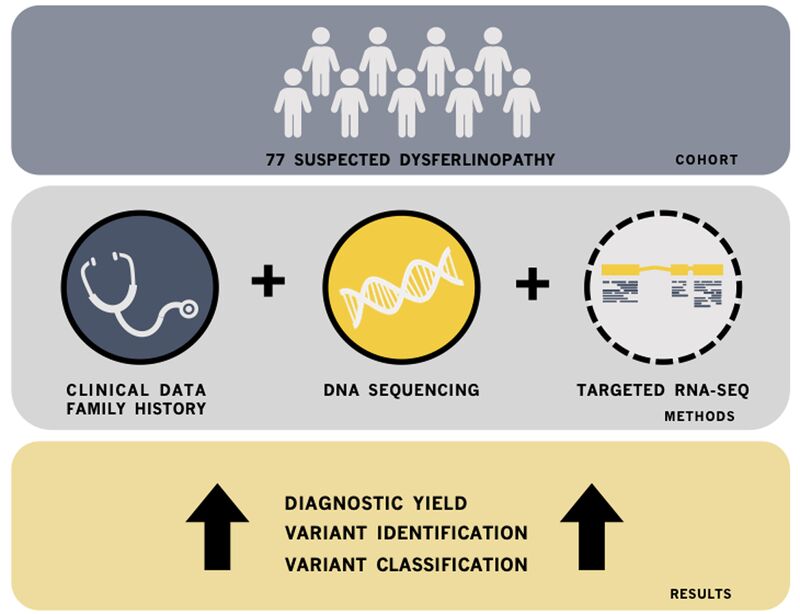
The Jain Foundation is excited to share that our research team has published a research article: Utilization of Targeted RNA-Seq for the Resolution of Variant Pathogenicity and Enhancement of Diagnostic Yield in Dysferlinopathy.
Our study harnesses the ability of RNA-Seq to assist in pathogenicity classification and diagnosis of dysferlinopathy. The fact that dysferlinopathy has significant clinical overlap between the 30+ subtypes of limb–girdle muscular dystrophy (LGMD) and that the DYSF gene contains a large number of variants of unknown significance (VUSs) ; can make obtaining a definitive diagnosis of dysferlinopathy challenging.
We performed targeted RNA-Seq using a custom gene-panel, as well as other DNA-based analyses (e.g., copy number variation analysis), in 77 individuals with a clinical/genetic suspicion of dysferlinopathy and evaluated the pathogenicity all 111 identified DYSF variants. The data from this study lead to a 48% increase in the diagnostic yield for this cohort of individuals and shows the incredible diagnostic power of RNA-Seq. Therefore, these types of analyses should be considered when typical DNA-based genetic analysis is not sufficient to provide a definitive diagnosis.
This work reflects the efforts of a dedicated team from Jain Foundation Inc, Georgia Tech Center for Integrative Genomics, Emory Department of Human Genetics, Emory Department of Pediatrics and PerkinElmer Genomics.